AI-guided protein engineering
Nature is pretty good at designing proteins. Scientists are even better. But artificial intelligence holds the promise of improving proteins many times over.
Medical applications for such “designer proteins” range from creating more precise antibodies for treating autoimmune conditions or cancers to more effective vaccines against viruses. Applications may extend beyond medicine to, for example, growing better crops that could be more nutritious or absorb more carbon dioxide from the atmosphere.
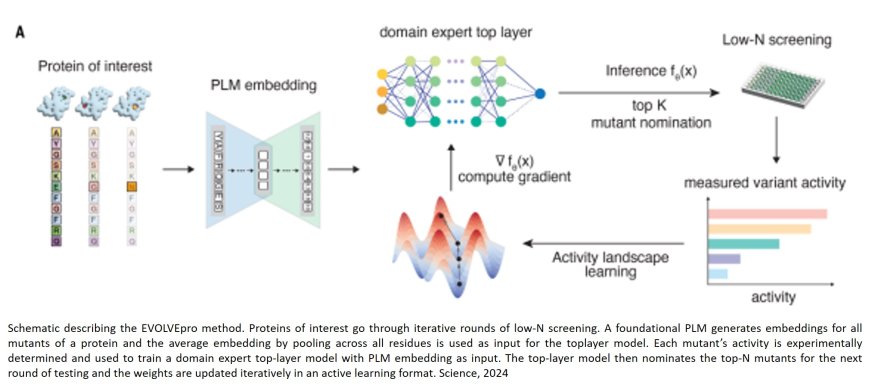
Investigators have developed an artificial intelligence (AI) tool known as EVOLVEpro that may represent a leap forward in protein engineering. In a paper published in Science, the research team demonstrates the tool’s ability to make proteins more stable, precise, and efficient by applying the model to engineer six proteins with different applications.
“The power of a tool like this is that we’re not restricted by evolution. Using AI, we can choose to optimize a protein to be better in whatever way is needed,” said co-senior author. “We can make a protein that’s better, faster, stronger. We can design it to be more efficient at binding to a target to improve a therapy or improve its function. If we can measure it, we can improve upon it.”
The concept of protein engineering is not new, but the emergence of AI and large language models is beginning to revolutionize the field. Protein language models (PLMs) can learn the “grammar” of proteins, reading protein sequences from across large genomic databases and offering suggestions that can improve proteins in ways that a scientist specifies. Much like new LLMs, EVOLVEpro acts as a layer on top of previous models, which can reason and provide more thought before responding.
“Protein modeling has advanced in recent years, and we wondered if we could now use large language models to essentially predict better protein sequences,” said co-senior author. “Our results consistently show how well this tool can work. We picked two clinically relevant antibodies—either already in use or close to human use—and found that with EVOLVEpro, we could engineer an antibody that could bind better and had better expression. Usually, you can do well on one of those outcomes, but here we saw improvements to both.”
The research team for the Science paper used EVOLVEpro to engineer six proteins. The researchers found that the two monoclonal antibodies EVOLVEpro engineered were up to 30-fold better at sticking to their target. A miniature CRISPR nuclease was five times more effective at making genetic changes. A protein used for n gene editing, called a prime editor, was twice as good at inserting sequences into different parts of the genome. A protein called Bxb1 integrase was four times more efficient at inserting DNA into cells for programmable gene integration applications, and a protein for RNA production, a T7 RNA polymerase, was 100 times better at making accurate copies of RNA, which is important for manufacturing mRNA for mRNA therapies or vaccines.
“We anticipate that this is just the beginning for EVOLVEpro, which will continue to improve with time and could be used for a wide variety of protein engineering applications,” said the author. “This technology marks the beginning of a new era where we can design proteins not just to match nature's designs, but to solve challenges nature never had to face—from creating more precise medicines to developing proteins that could help address global challenges like climate change and food security."