MetaFlowTrain to study ‘metabolic dialogue’ between microorganisms
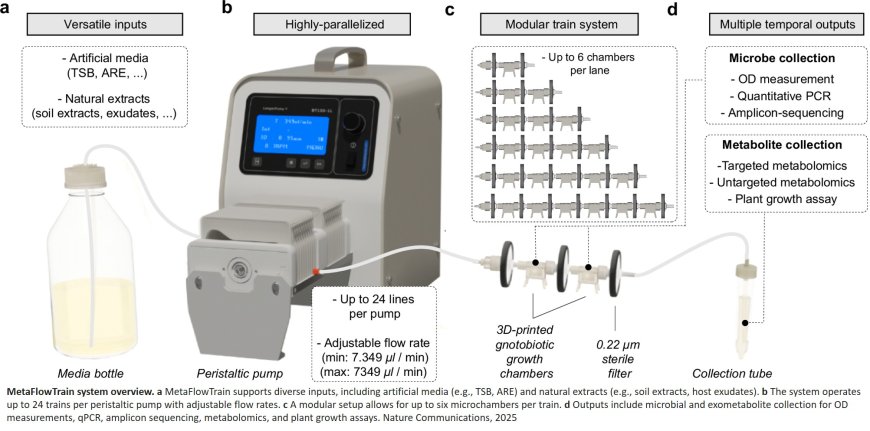
MetaFlowTrain: a highly parallelized and modular fluidic system for studying exometabolite-mediated inter-organismal interactions
Scientists have developed an innovative system – called MetaFlowTrain – that allows the study of metabolic exchange and interactions within microbial communities under different environmental conditions. The study is now published in Nature Communications.
Microbial communities consist of groups of microorganisms, such as bacteria, fungi, and other microscopic life forms, that coexist in specific environments. Although invisible to the naked eye, these communities are key for nutrient cycling, food web dynamics, and the degradation of pollutants in ecosystems. They are also essential for supporting the health of plants, animals, and humans by enhancing nutrient acquisition, bolstering the immune system, and protecting against harmful pathogens.
The tiny organisms within microbial communities interact both with their environment and with each other not only via physical contacts – but also through the exchange of metabolites. These so-called exometabolites are small molecules secreted by microorganisms in the environment, such as amino acids, organic acids, alcohols, and secondary metabolites. They play crucial roles in shaping microbial communities, influencing interactions through cooperation and competition. In these highly dynamic systems, it has been challenging for scientists to identify which microorganisms produce specific metabolites and how these metabolites affect other members of the microbial community.
The team worked on understanding the interactions between the members of plant-associated microbial communities. They realized that compartmentalizing the microorganisms would allow exclusion of the effects due to physical contact; thus, any remaining phenomena can be attributed solely to the exchange of metabolites and signals.
The fruit of this realization is the MetaFlowTrain, a purpose-built fluidic system that allows scientists to introduce different microorganisms into special 3D-printed microchambers. These microchambers are surrounded by filters that permit metabolite exchange while preventing microorganism transfer. These microchambers can stand alone or be put in a series (e.g., such as a train with different wagons), each holding different groups of microorganisms. For instance, with bacteria in the first chamber and fungi in the second, scientists can now observe how bacteria affect fungi – and vice versa – by simply switching the chambers.
The constant flow of fresh medium connecting these chambers allows different stress factors to be introduced to the system and prevents nutrient depletion in the microchambers. This represents a key innovation to identify novel microbial exometabolites with bioactive or signalling properties that shape microbial communities.
MetaFlowTrain is easy to build and affordable, and holds enormous potential for identifying molecules that mediate microbe-microbe interactions and microbe-host associations. Specifically, scientists can now better ‘eavesdrop’ on the metabolic dialogues that drive cooperation or competition in microbial communities. Further, harnessing the system's capabilities to discover novel antimicrobials that inhibit destructive plant pathogens presents new opportunities for sustainable agriculture and crop protection. In addition, the system could also be used to identify natural compounds for medicine and other fields.
“The staggering diversity of molecules produced by microbes is a result of millions of years of evolution. These molecules serve different functions that help these communities to survive, adapt, and thrive in various environments. Understanding the functions and the mode of actions of some of these molecules will drive agricultural innovations”, says the author.