Imaging-based STAMP technique for single-cell RNA analysis
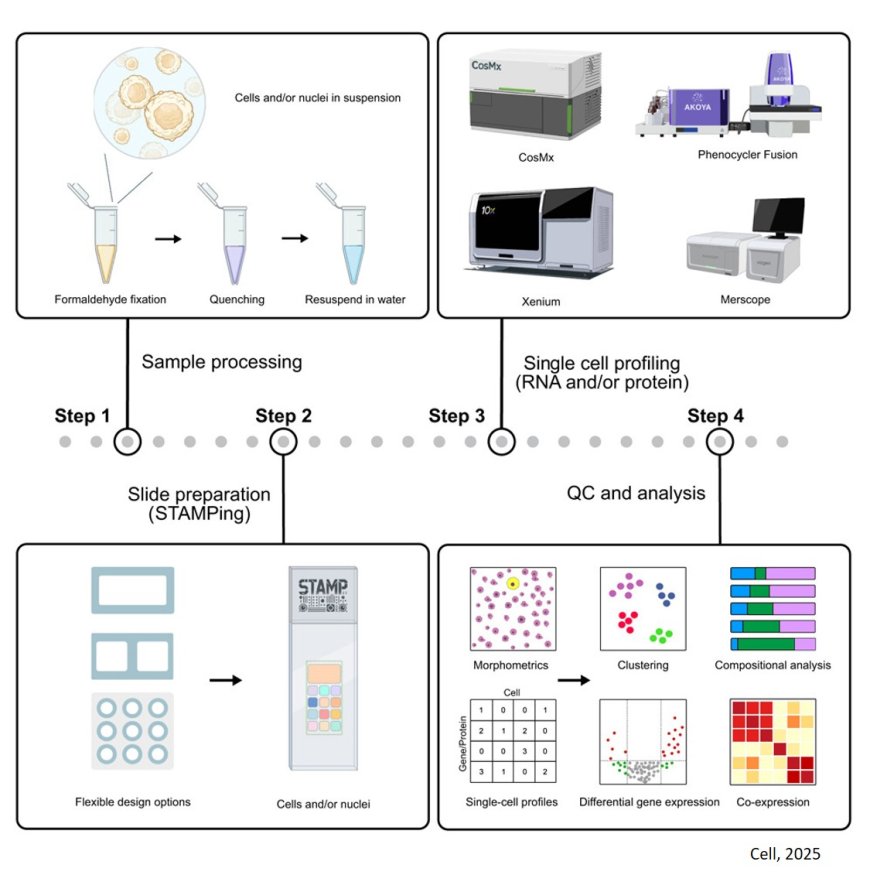
Single-cell RNA sequencing provides scientists with important information about gene expression in health and disease. However, the technique is expensive and often prohibits analysis of large numbers of cells. Scientists created a method that combines microscopy with single-cell RNA analysis to overcome these limitations. The technique called Single-Cell Transcriptomics Analysis and Multimodal Profiling through Imaging (STAMP) can look at millions of single cells for a fraction of the cost of existing approaches. The work was published in Cell.
“We’ve created a technique that gives us an advantage in the numbers game of single-cell analysis,” said a co-corresponding author. “It’s an order of magnitude more cost-effective and allows us to profile a million cells simultaneously, compared to tens of thousands typical of current methods, making it far more scalable.”
The researchers separated cells from tissues until they were individual, unconnected cells. They then fixed or “stamped” those cells onto microscopy slides, which kills the cells but “locks” them in time, including their shapes and expressed genes. The scientists then added molecules that light up under a microscope when bonded to specific RNA sequences.
The researchers compared their results to published atlases of gene expression data, finding they could characterize many immune cells simultaneously and discriminate between the developmental stages of induced pluripotent stem cells. They calculated that analyzing the immune cells from 1,000 individuals would normally cost $3.56 million while scaling STAMP to do the same analysis would cost $75,000, a 47-fold reduction.
“Current techniques for single-cell RNA sequencing require inference to determine things such as cell type, but STAMP allows us to directly examine the cells,” the author said. “Many current single-cell approaches also bias results based on cell shape, often missing irregularly shaped cells including neurons.”
Conventional RNA-sequencing techniques require cells to move through a tube before being analyzed. The more spherical and simpler the shape, the better the cell moves through these machines, causing some bias in results. The STAMP technique is performed on a microscope slide, avoiding this issue and allowing more diverse cell types to be represented.
To ensure sensitivity is also maintained, the researchers diluted cancer cells into a suspension of many other cells. STAMP detected two cancer cells within approximately 850,000 total cells on a single slide.
“Being able to profile a million cells is crucial because it only takes one cell to escape cancer treatment,” the author said. “We’ve shown STAMP allows us to visualize and detect these cells in a numerically advantageous and accessible way, an important feature for potential future development into clinical applications.”
Single-cell RNA sequencing remains prohibitively expensive, especially for those without access to the funding, specialized equipment or computational infrastructure it typically requires. However, almost all research institutions have easy access to microscopes.
“We as scientists believe what we can see,” the author said. “STAMP gives us the best of both worlds in single-cell analysis: quantitative gene expression data and the ability to visually examine the cells under a microscope. We hope that these features, combined with its accessibility and cost-effectiveness, enable others to discover new biology and clinical uses in the future.”