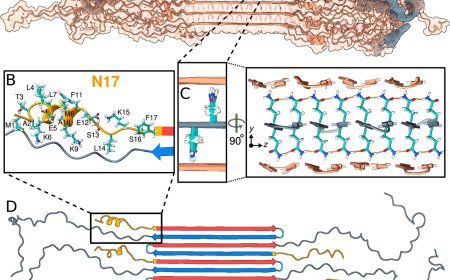
Structure of RNA self-folding!
The flexibility of RNA makes it notoriously challenging to study, as its structure can shift into numerous forms depending on environmental conditions. Traditional imaging methods, such as cryo-electron microscopy (cryo-EM) single-particle averaging (SPA) analysis, rely on averaging data from thousands of selected molecules with common shapes, making it difficult to capture the unique shapes of individual RNA molecules.
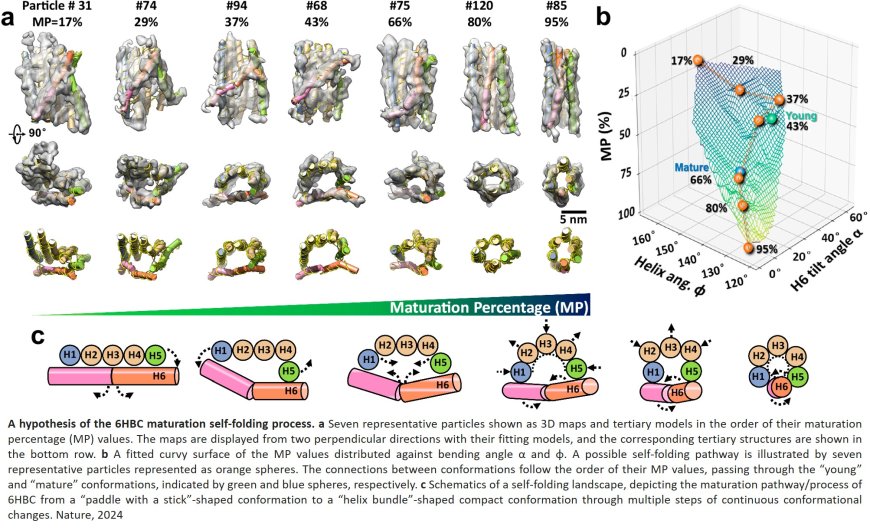
In a study published in Nature Communications, researchers explored the folding process of flexible RNA molecules. They employed an innovative technique capable of studying the 3D shape of individual molecules without averaging. This technique builds on advanced Individual-Particle cryo-Electron Tomography (IPET), a specialized approach that focuses on single molecule 3D imaging in cryo-preserved samples. Previously, this technique has been used to study how nucleosomes fold DNA and induce phase transitions.
Historically, scientists believed that obtaining 3D images from a single molecule was impossible due to weak signals. "It was considered a dead-end method since the 1970s," said the co-lead researcher.
In the current study, the researchers used IPET to study RNA origami – artificially structured RNA molecules engineered to fold into specific nanoscale shapes. The researchers had previously used the cryo-EM SPA method to study the 3D structure of RNA origami but the folding process remained elusive.
IPET allowed the researchers to capture a snapshot of RNA’s folding landscape through capturing molecules in various stages of folding, from immature states to their optimal shape. The researchers were able to observe a folding trap and a shift to a more compact form, enabling creation of a “movie” depicting RNA’s dynamic folding process.