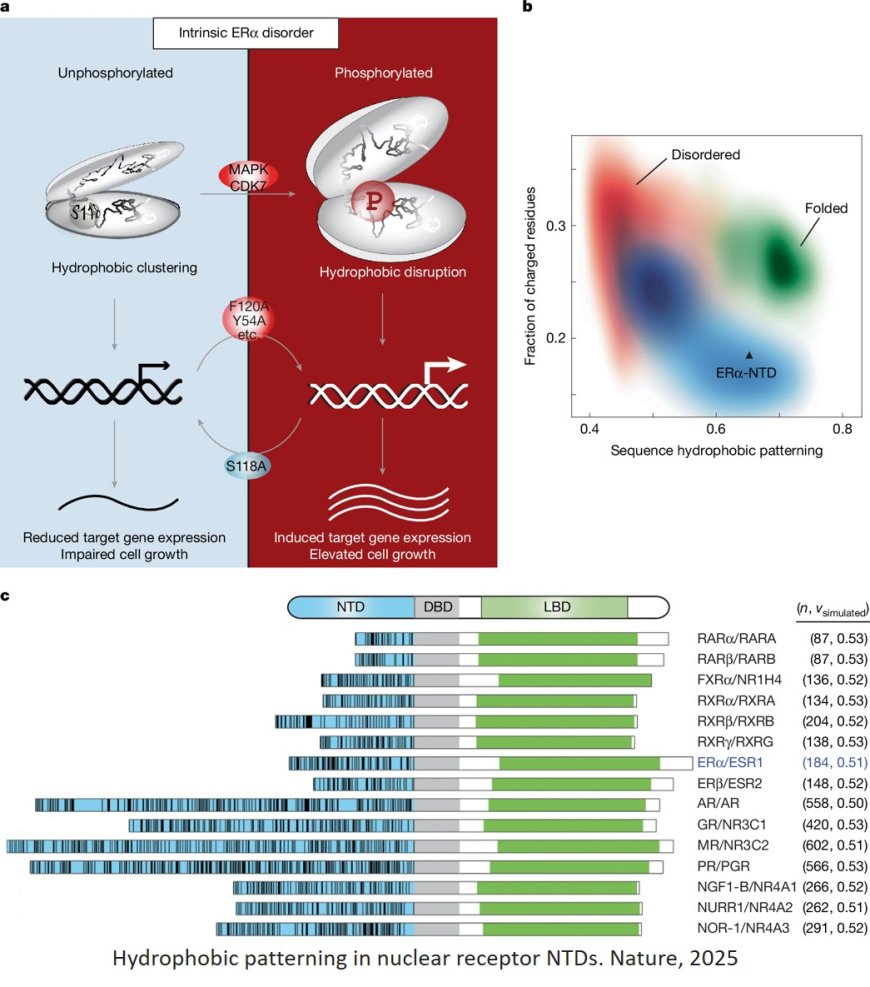
The sequence–structure–function relationship of intrinsic ERα disorder
Researchers have discovered how specific protein regions contribute to breast cancer.
Their study, published recently in Nature, focuses on what is known as the estrogen receptor, a protein linked in previous research to the development of roughly 70% of all breast tumors.
Just as a machine needs specific controls to function, proteins like the estrogen receptor control how cells grow and behave.
“We found previously unknown ‘molecular switches’ within the estrogen receptor that, while flexible, work together with remarkable precision to coordinate cellular processes,” said the senior author “Changing one part of the protein can trigger a chain reaction that affects how breast cancer cells grow.”
While more research is needed to possibly use their findings to develop new treatments, the discovery offers scientists new tools and ideas far beyond breast cancer, as similar protein regions are involved in many other diseases.
Breast cancer is the most commonly diagnosed cancer worldwide, with over 2.3 million new cases and 670,000 deaths reported in 2022, according to the World Health Organization. Despite the initial effectiveness of drugs that target the estrogen receptor, many patients eventually develop resistance, making therapies ineffective. This new discovery could result in more effective treatments and help explain why that might occur.
“Instead of simply shutting down the estrogen receptor,” the author said, “future drugs might be designed to target these newly discovered switches, offering new ways to control how the protein works.”
The research team used small-angle X-ray scattering and nuclear magnetic resonance spectroscopy tools to study the proteins switches in extraordinary detail.
They show that serine 118 phosphorylation triggers an unexpected expansion of the disordered domain and disrupts specific hydrophobic clustering between two aromatic-rich regions.
Mutations mimicking this disruption rescue ER transcriptional activity, target-gene expression and cell growth impaired by a phosphorylation-deficient S118A mutation.
These findings driven by hydrophobic interactions, extend beyond electrostatic models and provide mechanistic insights into intrinsically disordered proteins.