How DNA is recognized by proteins that control gene expression
With a new study in the journal Cell, researchers have contributed to increased knowledge about gene regulation in human cells.
How genes are turned on and off is crucial for the body’s cells to have different functions. In this process, certain proteins, so-called transcription factors, find and recognize different binding sites on DNA in the body’s cells to drive on or off signals. When this recognition goes wrong, it can give rise to many different types of diseases, such as cancer.
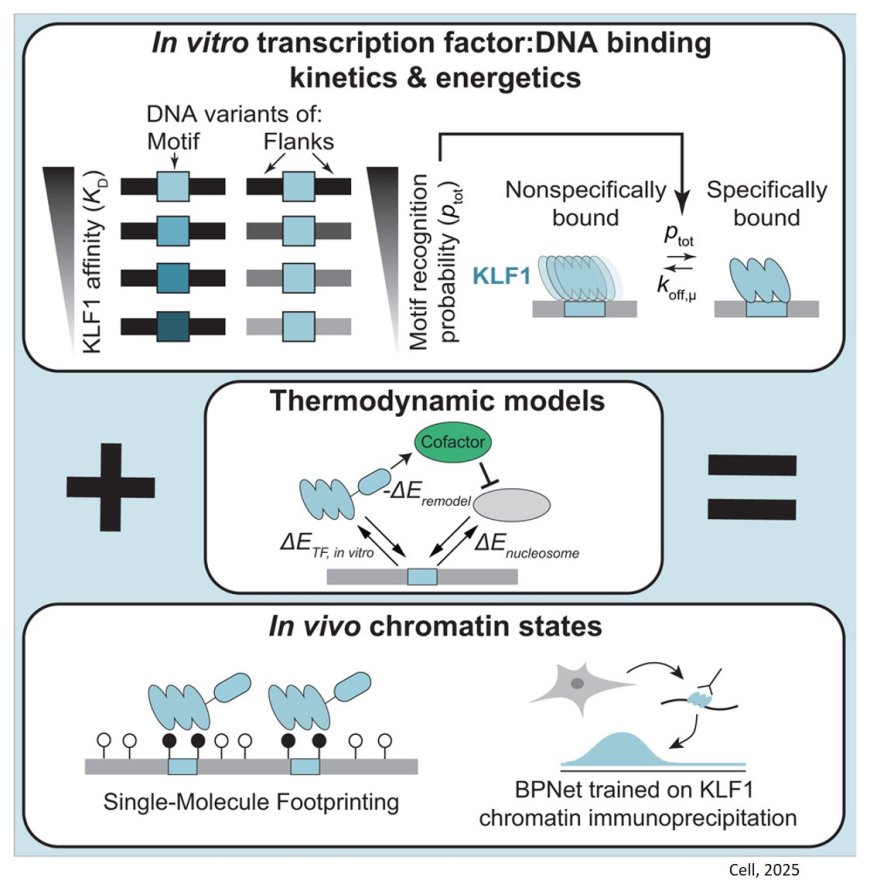
Now a group of researchers have studied in detail how DNA recognition works for one key transcription factor. This particular protein, known as KLF1, is essential for the development of red blood cells which carry oxygen around the body. In this study, they developed new experimental methods to make more precise measurements of how KLF1 interacts with DNA both in test tubes and in human cells.
“The most important result is that we show it is possible to understand the binding between this transcription factor and DNA in human cells, and that this behavior is consistent with what we measure in test tubes. That is an important basic science discovery”, says the author who has participated in the study that is now published in the journal Cell.
Transcription factor binding to DNA controls a lot in biology and causes the body’s cells to have different functions, explains the author:
“It is this process that controls that, for example, a nerve cell in the brain has different gene expression than a cell in the immune system. When the binding goes wrong, it can give rise to a great many different types of diseases. Genetic studies show that more than half of all mutations linked to traits such as genetic diseases occur in DNA sequence regions where transcription factors bind.”
First author emphasizes broader impacts from the study’s insights into how transcription factors read DNA:
“We discovered that this transcription factor pays attention to much more of the DNA sequence surrounding its binding sites than previously thought. By combining precise measurements in both test tubes and human cells with physical models, we were able to build a more complete picture of how DNA recognition works and, therefore, how gene regulation is encoded by DNA.”
The authors found 40-fold flanking sequence effects on affinity of transctiption factor binding. Motif recognition probability, rather than time in the bound state, drives affinity changes, and in vitro and in nuclei measurements exhibit consistent, minutes-long TF residence times.
https://www.cell.com/cell/fulltext/S0092-8674(25)01300-5
https://sciencemission.com/in-vitro-transcription-factor-affinities