RNA polymerase II at histone genes predicts outcome in human cancer
Using a new technology and computational method, researchers have uncovered a biomarker capable of accurately predicting outcomes in meningioma brain tumors and breast cancers.
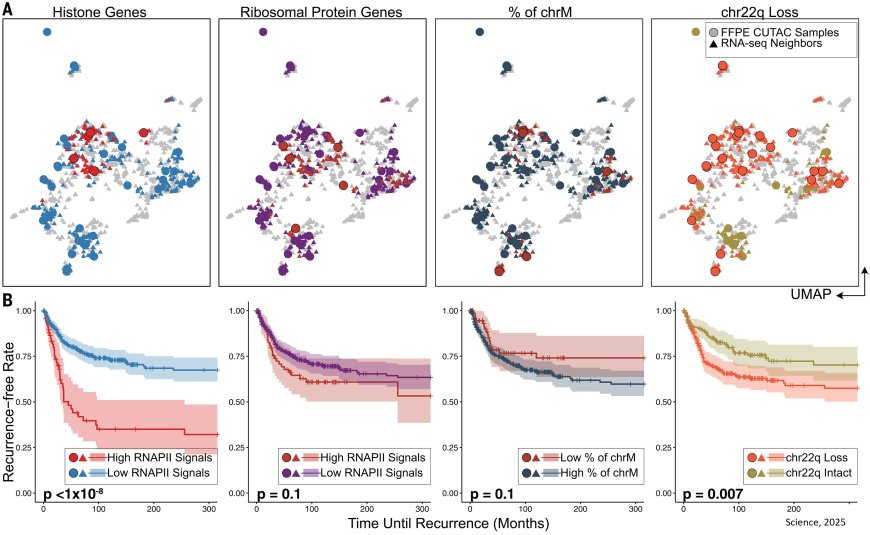
In the study, published in Science, the researchers discovered that the amount of a specific enzyme, RNA Polymerase II (RNAPII), found on histone genes was associated with tumor aggressiveness and recurrence. Hyper-elevated levels of RNAPII on these histone genes indicate cancer over-proliferation and potentially contribute to chromosomal changes. These findings point to the use of a new genomic technology as a potential cancer diagnostic and prognostic tool, which could improve precision oncology approaches.
“It has been overlooked that histone genes could be a rate-limiting factor in cell replication and, in turn, a strong indicator of tumor cell over-proliferation,” said a co-first author. “This is because current RNA sequencing methods are unable to detect histone RNAs due to their unique structure, meaning these libraries have vastly underestimated their presence. Our novel approach, combining a new experimental technology and computational pipeline, establishes a comprehensive ecosystem that can leverage biopsy samples from multiple cancer types to
Tissue biopsies are commonly stored for long-term use as FFPE samples, but the sample RNA becomes increasingly unstable over time due to degradation, leading to potentially lower-quality gene expression data.
The new technology – Cleavage Under Targeted Accessible Chromatin (CUTAC) – focuses on small, fragmented DNA non-coding sequences where RNAPII bind, located on the same chromosome as the gene they regulate, allowing scientists to directly measure gene transcription activity from the DNA.
When examining clinical samples using CUTAC technology across various cancer types, the researchers found that the expression of histone genes was consistently and significantly higher in tumor samples compared to normal tissue samples.
Histone proteins provide essential structural support for DNA in chromosomes, acting as spools around which DNA strands wrap. These proteins have been well studied, but most current tools to study gene expression rely on RNA sequencing. Histone RNA is unique in that its structure prevents the RNA molecules from being detected by current methods.
Thus, the expression of histone genes may be significantly underestimated in tumor samples. The researchers hypothesized that the increased proliferation of cancer cells leads to a very elevated expression, or hypertranscription, of histones to meet the added demands of cell replication and division.
In tumor samples, the RNAPII enzyme signals found on histone genes were reliably able to distinguish between cancer and normal samples.
RNAPII signals on histone genes also correlated with clinical grades in meningiomas, accurately predicting rapid recurrence as well as the tendency of whole-arm chromosome losses. Using this technology on breast tumor FFPE samples from 13 patients with invasive breast cancer also predicted cancer aggressiveness.
“The technique we developed to examine preserved tumor samples now reveals a previously overlooked mechanism of cancer aggressiveness,” said the senior author. “Identifying this mechanism suggests it could be a new test to diagnose cancers and possibly treat them.”