Slow editing of protein blueprints leads to cell death
Genes contain the essential building instructions for life, guiding cells on which amino acids to assemble in what sequence to produce specific proteins. The human genome codes for about 20,000 such instructions. “Nevertheless, our cells can produce several hundred thousand different proteins,” explains the author.
This diversity is enabled by a process known as “splicing.” When a cell requires a protein, it generates a copy of the relevant instructions in the cell nucleus. During splicing, this transcript undergoes modification: a cellular editing complex, the spliceosome, removes certain segments. The outcome varies depending on which parts are cut out, resulting in distinct blueprints for different proteins.
This process is crucial for the life of the cell. “The spliceosome is composed of multiple components that secure production of functional proteins controlling cellular life,” explains the author. “If this complex is disrupted, it can lead to the death of the affected cell. For this reason, spliceosome inhibitors are considered as potential anti-cancer drugs.” However, the downside is that a complete blockade of this "editing office" also affects healthy cells, resulting in significant side effects of any spliceosome inhibitor developed so far.
In an international study, researchers have now identified a mechanism that interferes with the splicing process in a more subtle way. It is related to a specific part of the spliceosome, composed of three subunits known as U4/U6.U5.
“We already knew that certain mutations in these subunits are linked to the eye disease retinitis pigmentosa,” says the first author of the study. “What we didn’t yet understand was the exact impact of these mutations.”
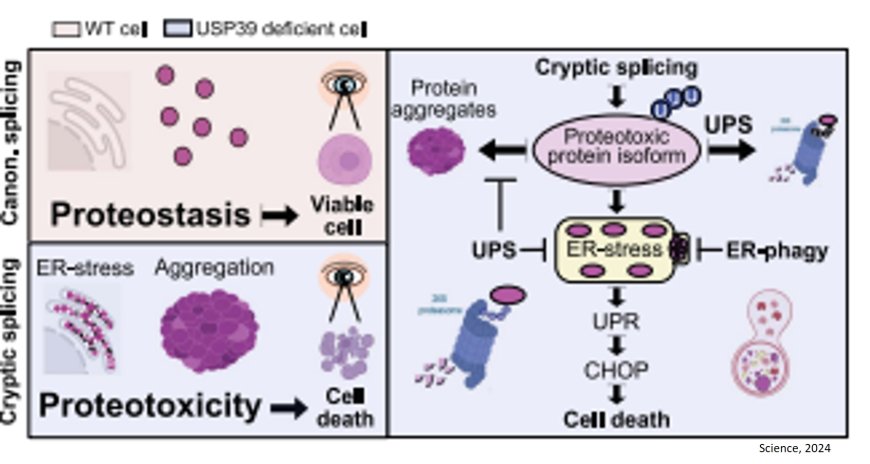
In experiments with zebrafish, the team has now managed to fill this knowledge gap. Their findings reveal that spliceosome subunits U4, U5 and U6 are normally stabilized as a complex by a protein called USP39. However, when subunits are mutated or USP39 is absent, the stability of the tripartite complex is compromised, causing the spliceosome to lose precision. During splicing, U4/U6.U5 normally ensures the immediate and correct re-joining of loose ends after a transcript has been cut. Without USP39, or when subunits are mutated, this re-joining is delayed.
“This increases the likelihood of incorrect connections, as we were able to show in computer simulations,” explains the author. This results in incorrectly edited transcripts, on the basis of which the cell then produces dysfunctional proteins. These accumulate and can form aggregates inside the cell. Cells have a waste disposal system to clear out defective molecules, and this protective mechanism was activated in cells lacking USP39. Over time, however, this "garbage disposal" became overwhelmed by the protein aggregates, leading to cell death in the zebrafish retina.
“The discovery of this mechanism was unexpected,” emphasizes the author. “We suspect it may also explain why retinal cells in retinitis pigmentosa patients die. Defective splicing variants might also play a role in the development of neurodegenerative diseases like Alzheimer’s or Parkinson’s. On the other hand, this mechanism may be targeted by new therapeutic approaches for types of cancer that are highly dependent on the correct function of the spliceosome.”
Some highly aggressive tumors produce large amounts of USP39 and related splicing factors, likely due to their high division rate: To maintain constant protein production, they require highly precise splicing, a function that USP39 provides. “Blocking USP39 in these cancer cells could selectively kill them,” the author explains. “Healthy cells, on the other hand, with their much lower division activity, would be spared. This is an approach that we are currently investigating.”