Identification of antimalarial target binding partner
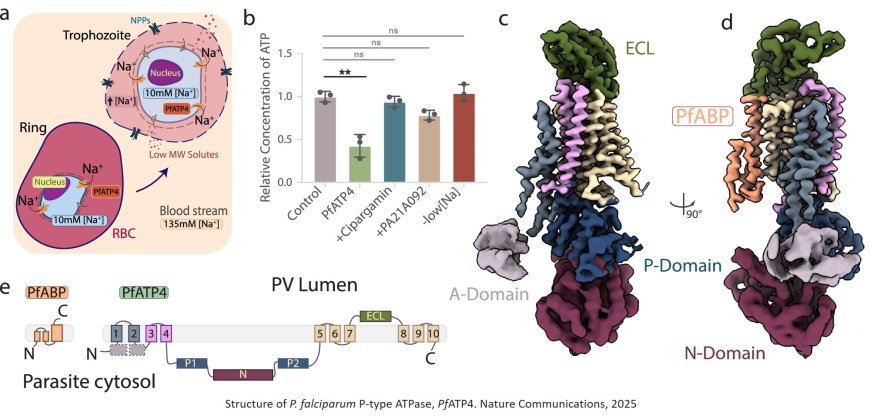
As malaria parasites around the world become resistant to anti-malarial drugs and progress against the disease stalls, researchers are looking for new compounds to kill the parasites. A sodium pump essential to the parasite’s survival, PfATP4, has emerged as one of the most attractive drug targets, but parasites have rapidly developed resistance to experimental PfATP4 inhibitors.
The new study provides researchers with critical information about PfATP4 that could bring about more durable inhibitors. The study presents the first high-resolution 3D structure of PfATP4—revealing new sites where inhibitors could bind—and identifies a previously unknown but essential binding partner, opening an entirely new avenue for anti-malarial drug development.
“Every year, malaria parasites adapt to outsmart our medicines. With the structure of PfATP4 now in hand and the discovery of this unknown partner, we have identified vulnerabilities that can be exploited for new therapies,” says a co-senior author.
The new research underlines the importance of studying the malaria parasite in its natural host cell. Previous attempts to elucidate the 3D structure of PfATP4 have followed standard procedures for structural studies: Researchers first insert a gene into a yeast or bacteria cell, culture the cells to create enough copies of the gene’s protein for analysis, and then obtain images of the proteins using cryo electron microscopy.
But expressing P. falciparum genes in cells other than the parasite’s natural hosts often fails.
Several years ago, the researchers pioneered techniques that allow researchers to obtain high-resolution 3D structures of P. falciparum proteins isolated directly from parasite-infected blood cells.
In the new study, the research team used innovative techniques to successfully visualize the PfATP4 sodium pump, allowing the scientists to precisely map where clinically relevant resistance mutations occur.
“The findings provide a blueprint for next-generation drug discovery, offering ways to design molecules that target PfATP4,” says the other co-senior author.
Unexpectedly, the images also revealed a previously unknown protein bound to the pump. The new protein, which the researchers named PfATP4 Binding Protein (PfABP), appears to stabilize and regulate PfATP4’s function and is essential for parasite survival.
“We found that loss of PfABP led to the rapid degradation of the PfATP4 sodium pump and death of the parasite,” says co-first author. “Because PfABP seems to be less prone to mutations, drugs that target it may be harder for parasites to evade.”
The senior author says that the discovery of the binding protein would not have been possible without techniques that allow the parasite’s proteins to be studied in their natural state.
“These endogenous methods allow us to understand proteins as they exist in their cellular milieu and enable the discovery of completely new biology,” the author says.