Platform for in vitro production of human microglia cells from iPSC
Microglia are a specialized type of immune cells that accounts for about 10% of all cells within the brain and spinal cord. They function by eliminating infectious microbes, dead cells, and aggregated proteins, as well as soluble antigens that may endanger the brain and, during development, also help shape neural circuits enabling specific brain functions.
When microglia don’t function properly, they can trigger neuroinflammation and fail to clear away damaged cells and harmful protein clumps – such as the neurofibrillary tangles and amyloid plaques seen in Alzheimer’s disease. This contributes to numerous neurodegenerative diseases, including Alzheimer’s, Parkinson’s and Huntington’s disease, as well as Amyotrophic lateral sclerosis (ALS), Multiple sclerosis, and other disorders. In fact, neuroinflammation can occur even before proteins start to form pathogenic aggregates and, in turn, accelerates protein aggregation.
Researchers and drug developers aiming to better understand and target microglia functions in the brain are challenged by the fact that human microglia can only be obtained through biopsies, and rodents’ microglia differ from their human counterparts in many critical features. This supply issue prompted them to work on methods to create microglia in the culture dish using stem cells as a starting point. However, to date, this process has remained inefficient, and requires weeks to complete at significant costs.
Now, a research team has devised a solution for creating microglia with strong functional similarities to human microglia from induced pluripotent stem cells (iPSCs) within four days, compared to 35 days it takes to obtain similar, yet less fine-tuned cells in a conventional differentiation process. Their approach builds on a previously developed technology known as “TFomeTM” that can be used to drive multiple cell differentiation processes in the dish more efficiently than other methods can. In TFomeTM technology, critical instructive proteins known as transcription factors (TFs) that orchestrate entire gene expression programs are expressed in iPSCs to specify their fate toward differentiated functional cell types.
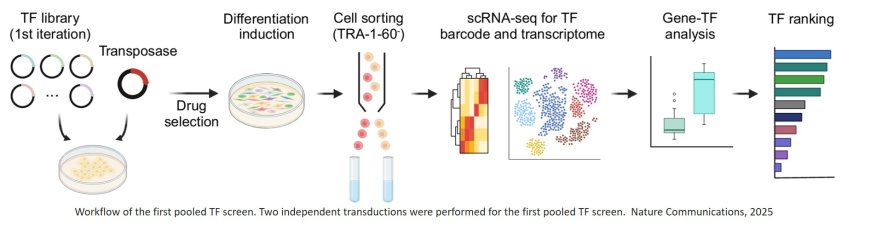
In their new study, the team designed microglia-specific libraries of TFs, and then performed iterative rounds of screening distinct combinations of TFs for their ability to convert iPSCs to microglia-like cells. To investigate individual resulting cells, they used single cell RNA sequencing (scRNA-seq) technology to determine how similar their gene expression had become to that of actual microglia. Through this process, they identified a potent cocktail of six TFs enabling the ultra-fast production of microglia-like cells. Their findings are published in Nature Communications.
“We advance transcription factor libraries as a platform technology here in a synthetic biology paradigm. By integrating it with single-cell RNA data-driven analysis and iterative rounds of optimization, we succeeded in creating much sought-after human microglia cells in the dish,” said the author. “This cell differentiation approach can open many avenues of brain disease-focused research and new therapeutic perspectives. Equally relevant, it can be applied to the generation of other hard-to-get and therapeutically relevant cell types that require complex transcriptional scenarios.”
In 2021, co-authors had created a comprehensive library of 1,732 human TFs and their variants, a vital piece of the TFomeTM platform technology, and identified individual TFs with the ability to generate specific cell types for potential use in the accelerated manufacturing of next-generation cell therapies.
“To create human microglia cells in vitro, using the TFomeTM process, we realized that we don’t have to screen the entire library but, based on a large body of earlier developmental and disease studies, could start by making a smart choice,” said first-author. “So, we came up with a collection of 40 TFs whose induced gene expression profiles were typical for primary human microglia and devised a strategy to express random combinations of five to seven of them in single iPSCs.”
To determine which TF combinations most effectively induced the gene expression of differentiating microglia, the researchers applied statistical and computational methods that enabled the team to glean microglia-like gene expression changes in scRNA-seq data they acquired from thousands of single cells following only a few days of culture, and then rank the corresponding TF combinations. The result of this first round of screening were three TFs (SPI1, CEBPA, and FLI1) that, together, turned on a microglia-specific differentiation program in iPSCs.
Although the cells exhibited desired microglia-like transcriptional and morphological changes, the team realized that they still had not reached the functional maturity of actual primary human microglia. “We argued that we could go through iterative rounds of this design-screen-validate cycle, meaning that adding new TFs in consecutive rounds could improve outcomes and lead to more superior TF combinations,” said the author.
By looking for additional TFs that were still expressed at lower levels in differentiating iPSCs that received the three-TF cocktail, compared to primary human microglia, and through different computational predictions, the researchers compiled a second set of 42 additional TFs. Their sc-RNA-seq and computational analysis pinpointed another three among them (MEF2C, CEBPB, and IRF8) that, in an expanded six-TF cocktail, advanced microglia differentiation even further.
The team tested whether the resulting cells, like actual microglia, could be activated by stimuli typical of brain infections and neurodegenerative diseases. They found that the cytokine interferon gamma (IFNg),whose increase in the presence of infectious pathogens, induced a microglia-specific gene expression pattern in the differentiated cells. Also, the so-called TDP-43 protein, which forms aggregates in ALS patients, caused human microglia-like gene expression changes. “Building on this proof-of-concept study, we believe that by identifying additional TFs, developing ways to further fine-tune the expression strength of individual TFs, and order of their appearance on the four-day time scale, we can further hone specific microglia identities and even create microglia subtypes with dedicated functions in the brain,” said the author.
https://www.nature.com/articles/s41467-025-59596-3
https://sciencemission.com/microglia-like-cells-from-human-iPSC