Platform for annotating and interpreting single-cell populations
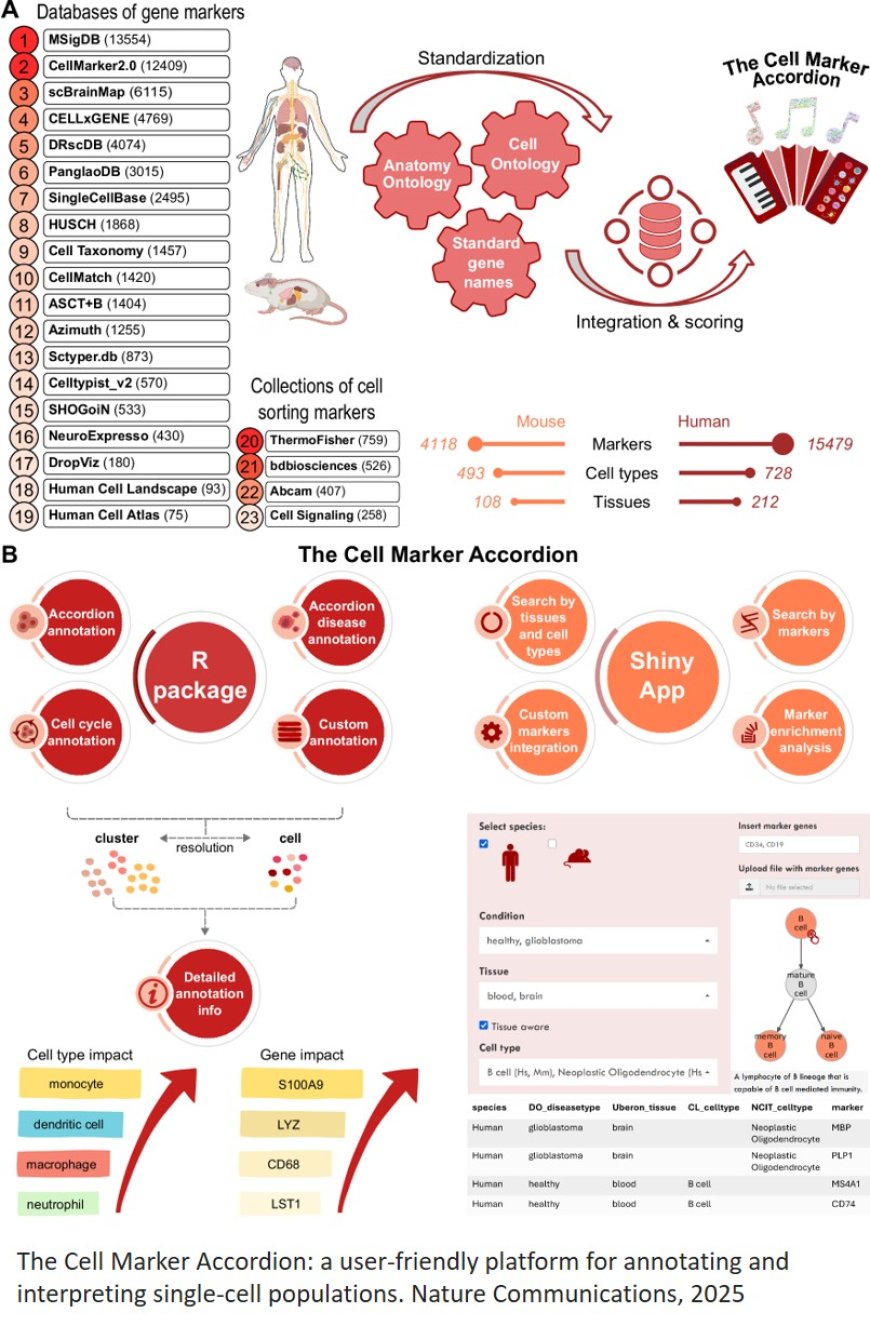
"With Cell Marker Accordion we wanted to build a tool that helps researchers not only to classify cells, but also to understand why they have been classified in a certain way", explains the first author of the work. "Often software give a result, but do not say how they got there. We wanted to do something more transparent and useful for people working in clinical settings."
The name of the instrument – "Accordion" – recalls the idea of harmonizing different data to provide a more robust result.
The software has been designed to help identify cell types in biological samples both under normal conditions and in the presence of disease. It can, for example, indicate the presence of leukemic stem cells or tumour plasma cells, also suggesting which genes could be involved in the alterations.
"Our tool does not limit itself to indicating what type of cell is present, but also helps to find out which genes make that cell unique and different from the others," adds the corresponding author of the research. "This can help identify new biomarkers or therapeutic targets."
One of its strengths is accessibility. In addition to the software package for those with bioinformatics skills, the Accordion has a web version with an intuitive interface that can easily be used even by non-programmers.
One of the future goals of the project is to adapt the instrument to new types of data and keep it updated over time, to make sure that the scientific community can always count on a reliable tool. "A scientific software does not end with a publication," concludes the author. "quite the contrary: it must be maintained, constantly improved, made more and more useful in line with new discoveries. This too is a service to research."