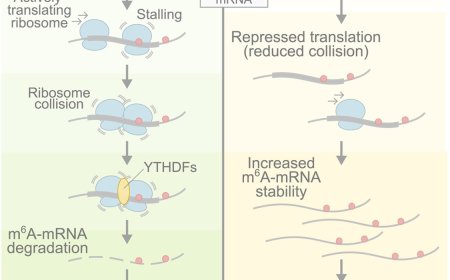
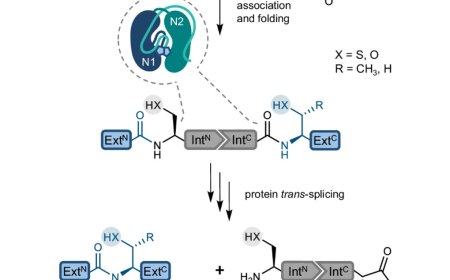
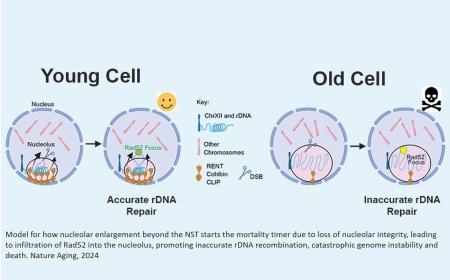
Synthetic lethal interactions among DNA repair factors
The DNA of human cells consists of a sequence of about 3.1 billion building blocks. Cells go to great lengths to maintain the integrity of this vast store of information. They constantly untangle knots in the DNA strand and create new chemical bonds when a strand of DNA breaks somewhere in the nucleus.
“When people read about repairing genetic material, they often think of it being in response to exposure to toxins or radiation,” says the senior author. However, repair mechanisms not only defend against external threats; they also play a crucial role in helping cells survive the challenges they face in their daily fight for survival.
Scientists have long known that more than 500 genes (out of approximately 20,000 human protein-coding genes) are crucial for DNA repair. By comprehensively analysing the interactions of these genes, the team have gained important new insights into how cells maintain the integrity of their genome. The researchers have discovered a wealth of new interactions and have identified potential new targets for cancer therapy, as reported in the journal Nature.
The researchers modified human cells in culture to switch off two of the repair genes simultaneously. “We took a systematic approach and looked at all possible combinations,” says the author. That’s easier said than done, because the researchers looked at nearly 150,000 different combinations of inactivations. “There was a lot to figure out,” say the two lead authors who carried out most of the study.
Inactivating a single gene is usually not enough to produce a noticeable effect because another gene often compensates for the missing function. “Human cells love redundancy,” says the senior author. Only when the backup is also switched off do they lose the ability to repair the genetic material. Sooner or later, the damage accumulates to a point where survival is no longer possible.
This was the case for about 5,000 inactivated gene pairs. The researchers carried out CRISPR interference (CRISPRi) screening to comprehensively map the genetic interactions required for survival during normal human cell homeostasis across all core DNA damage response genes. In their paper, the researchers provide a detailed description of the molecular interactions lost for two specific gene pairs. In their investigations, they uncovered previously unknown links that appear to be essential for the cell’s survival.
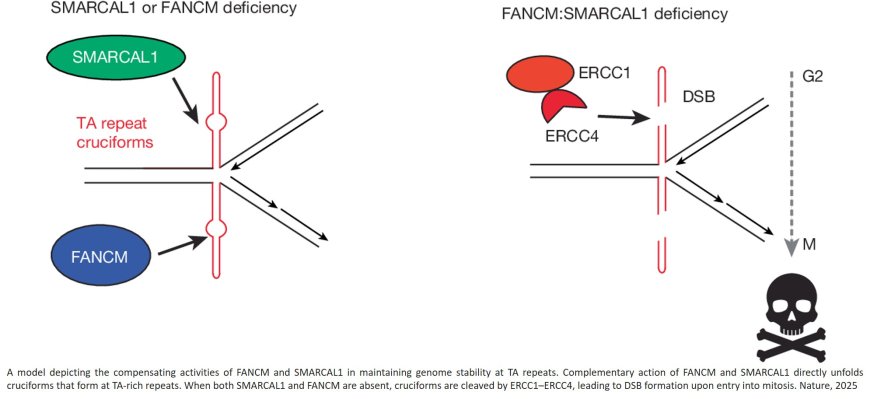
They found that WDR48 works with USP1 to restrain PCNA degradation in FEN1/LIG1-deficient cells. They also found that SMARCAL1 and FANCM directly unwind TA-rich DNA cruciforms, preventing catastrophic chromosome breakage by the ERCC1–ERCC4 complex.
The researchers’ work not only offers important insights but also provides new possibilities for cancer therapy. Cancer cells have more mutations than normal cells. For this reason, some of the 500+ repair genes are already switched off in some cancer cells. “Our research has identified the additional genes that need to be inactivated to prevent the growth of cancer cells,” says the author.
In the paper, the team lists a number of previously unknown links between common cancer mutations and molecular targets that can be blocked with drugs. “These newly discovered potential vulnerabilities in cancer cells now need to be tested. We have shown the pathways through the dark forest,” says the author. “Now it’s easy to walk those paths.” The team have created a new web platform where their results are publicly available. “We hope that other researchers will benefit from this and actively use the platform,” adds the author. This would help to ensure that new knowledge about genetic interactions bears fruit as quickly as possible.