Visualization of real-time protein translocation across the bacterial membrane
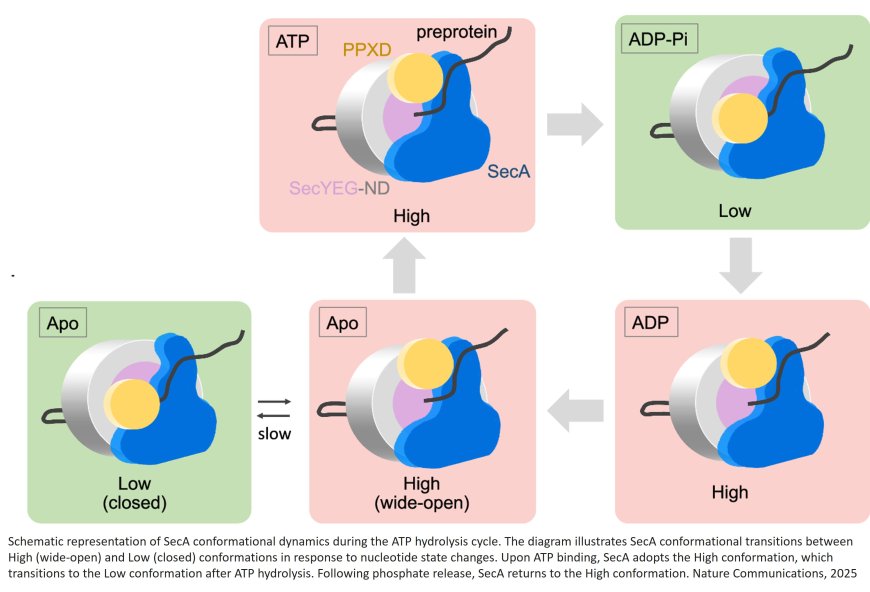
Protein translocation is an essential, nano-scale dynamic process that facilitates the movement of proteins across cellular membranes, enabling them to reach specific locations within the cell or to be transported outside the cell. This process occurs through membrane protein complexes that provide necessary channels for the movement of proteins. In bacteria, a group of proteins forms the SecYEG-SecA complex, which helps cellular proteins translocate across the cytoplasmic membrane. The SecYEG component is a channel through which SecA drives protein translocation, using energy from a molecule called adenosine triphosphate (ATP), also known as the "energy currency" of the cell. Despite many different approaches, observing this process in detail has been quite challenging.
For the first time ever, researchers directly visualized protein translocation across membranes using high-speed atomic force microscopy (HS-AFM)—an event that had been biochemically predicted but never observed. This pioneering study, published in Nature Communications; the researchers successfully visualized how the SecYEG-SecA complex helps mediate the translocation of unfolded proteins across the bacterial membrane at a molecular level.
The researchers specifically focused on the conformational changes in SecA during the ATP hydrolysis cycle, which is critical to the translocation mechanism. The hydrolysis of ATP serves as the energy source that drives protein transport.
By analyzing the SecYEG-SecA complex dynamics using HS-AFM, the team captured real-time snapshots of SecA transitioning between two distinct conformational states due to the alteration of a region of SecA, PPXD—referred to as the "High" and "Low" states. These changes were linked to the ATP hydrolysis cycle.
“Thirteen years ago, we embarked on this journey to visualize protein translocation across membranes. The challenge of achieving the necessary spatiotemporal resolution pushed the very limits of high-speed AFM. Yet, through relentless dedication, meticulous sample preparation, and patient observation, we are thrilled to have captured these truly groundbreaking images,” shares the author.
Detailed analyses also estimated the rate of protein translocation to be about 2.2 amino acid residues per second, and a single SecYEG-SecA complex was sufficient for successful protein translocation.
“We not only succeeded in capturing the protein membrane translocation process in real-time—an event previously elusive to visual observation—but also estimated the structural changes of motor proteins and their transport speeds, marking a significant breakthrough. Moving forward, we plan to apply this method to analyze other membrane proteins. This study paves the way for future research in visualizing these dynamics, potentially leading to better comprehension of the molecular mechanisms behind protein translocation,” concludes the author.