AI-based software maps proteins and lipids within cells
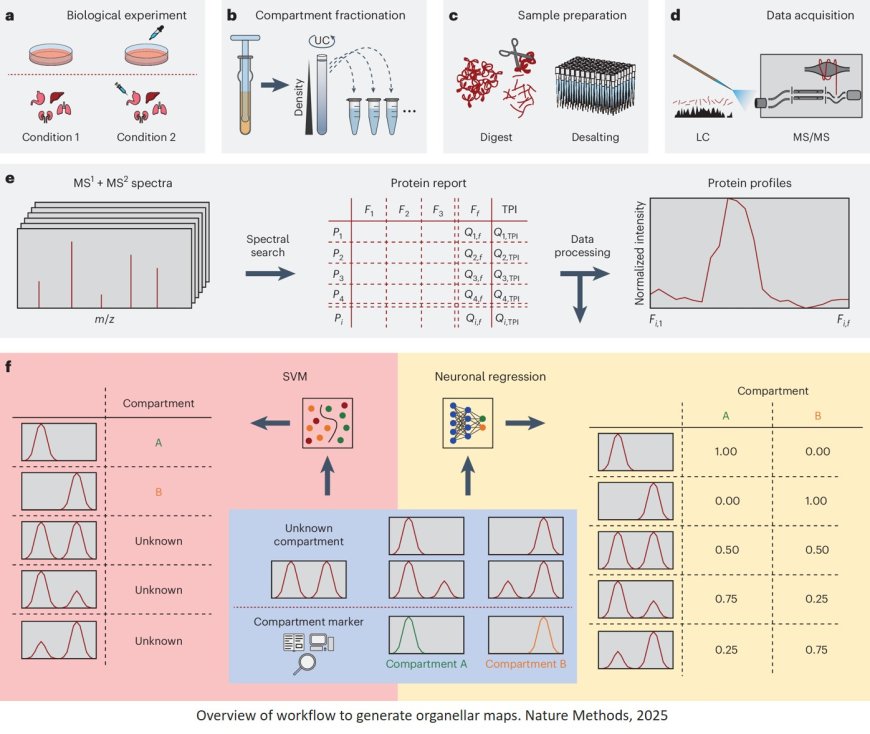
Existing tools for spatial proteomics often have constraints. Many are not equipped to predict multiple localizations for individual proteins or to quantify across different cellular compartments. In addition, their use frequently requires programming knowledge and lacks accessible interfaces, which can limit broader application. Spatial lipidomics has remained challenging due to the absence of reliable markers for lipid localization.
C-COMPASS was developed to address these methodological gaps. The software uses neural networks to predict multiple subcellular protein localizations and incorporates total proteome data to assess changes in protein distribution and organelle abundance. It includes a graphical user interface and standardized processing steps designed to support reproducible analyses.
“With C-COMPASS, we wanted to create a tool that makes spatial proteomics more accessible and easier to reproduce,” says the author. Project leader adds: “For the first time, it also allows us to explore spatial lipidomics by combining proteome and lipidome data in a unified workflow. We can now generate cellular atlases of organs and tissues at combined proteome and lipidome levels, what enables researchers to address many new questions.”
The research team applied C-COMPASS to investigate spatial protein distributions in humanized liver tissue and examined how these patterns shift under different metabolic conditions. They then extended the workflow by integrating proteomic and lipidomic data, enabling spatial lipidomics for the first time. To localize lipids, the researchers mapped them onto spatial reference maps derived from proteomics data. This approach was applied to humanized mouse liver samples and revealed changes in lipid distribution associated with metabolic perturbations.
The team plans to apply C-COMPASS to a variety of datasets to gain deeper insights into dynamic, metabolism-related changes in protein localization. They are also working to improve the software further – with features like support for other spatial omics methods, such as spatial transcriptomics.