New mechanisms for bacterial motility and DNA transfer between bacteria decoded
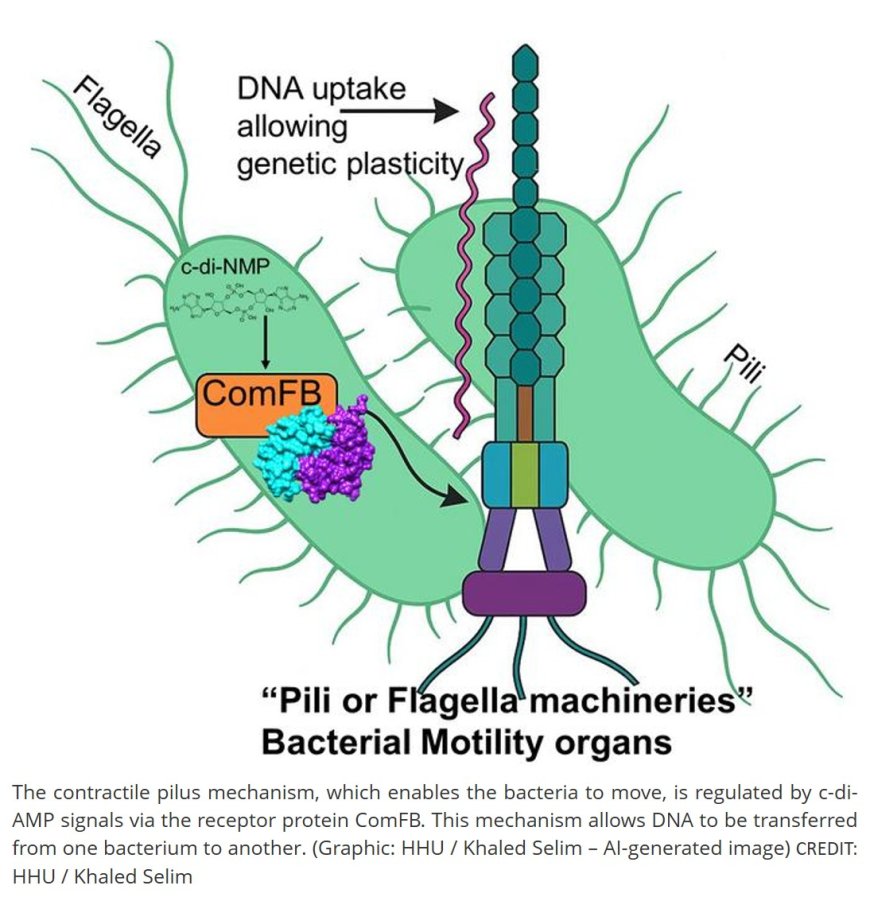
Bacteria are constantly moving by help of motility organs called flagella or pili to colonize new niches. Also, bacteria can exchange information, like “speaking to each other”, and thus acquire new abilities through the exchange of DNA materials. These motility organs play important roles in DNA uptake to exchange genetic information between different bacteria, allowing what’s so-called genomic plasticity. Therefore, bacterial motility organs contribute to bacterial pathogenicity, colonizing hosts, biofilm formation and spreading of antibiotics resistance.
Scientists have discovered a new family of signaling proteins, widespread in bacterial kingdom and contributes to regulating bacterial motility and DNA uptake mechanisms. The researchers reported their finding in two publications in the journals Cell Discovery and Proceedings of the National Academy of Sciences (PNAS).
In these two studies, the research team revealed that the second messenger molecules (c-di-GMP and c-di-AMP), together with the newly discovered ComFB receptor protein, regulates both bacterial motility and DNA uptake, therefore possibly contributing to bacterial pathogenicity. The first author of both publications: “The ability of different ComFB proteins from various bacteria to bind and precisely integrate the motility/DNA uptake signal(s) is reported by the second messenger di-nucleotides (c-di-GMP and c-di-AMP).” These characteristics were found in cyanobacteria, Bacillus subtilis and Vibrio cholerae, the causative agent of Cholera outbreak.
The senior author: "c-di-AMP belongs to one the relatively newly discovered class of 'cyclic dinucleotide-type second messengers,' whose cellular functions are not yet fully understood. In these studies, we show that the ComFB proteins sense c-di-AMP and additionally c-di-GMP are essential for regulating the natural competence and bacterial motility." The term "natural competence" describes the ability of bacterial cells to uptake DNA molecules and integrate them into their own genome to exchange the genetic information between each other. This process, for example, contributes mainly for spreading antibiotic resistance from initially only a few bacteria to entire populations, and also across different species boundaries.
It is still unclear which other groups of bacteria—besides the bacteria studied—also use this mechanism. “If pathogenic bacteria with clinical relevance also use it, this could pave the way for new strategies to fight multi-resistant bacteria,” the author emphasizes.