Neurotransmitter classification from electron microscopy images
Like many good ideas in science, it started with a walk in the woods.
During a stroll through the Berlin Botanic Garden in 2019, the researchers started chatting about a familiar topic: how to get more information out of insect connectomes.
These wiring diagrams give researchers unprecedented information about brain cells and how they connect to each other, but they don’t tell scientists how the signal from one neuron affects the other neurons in its network.
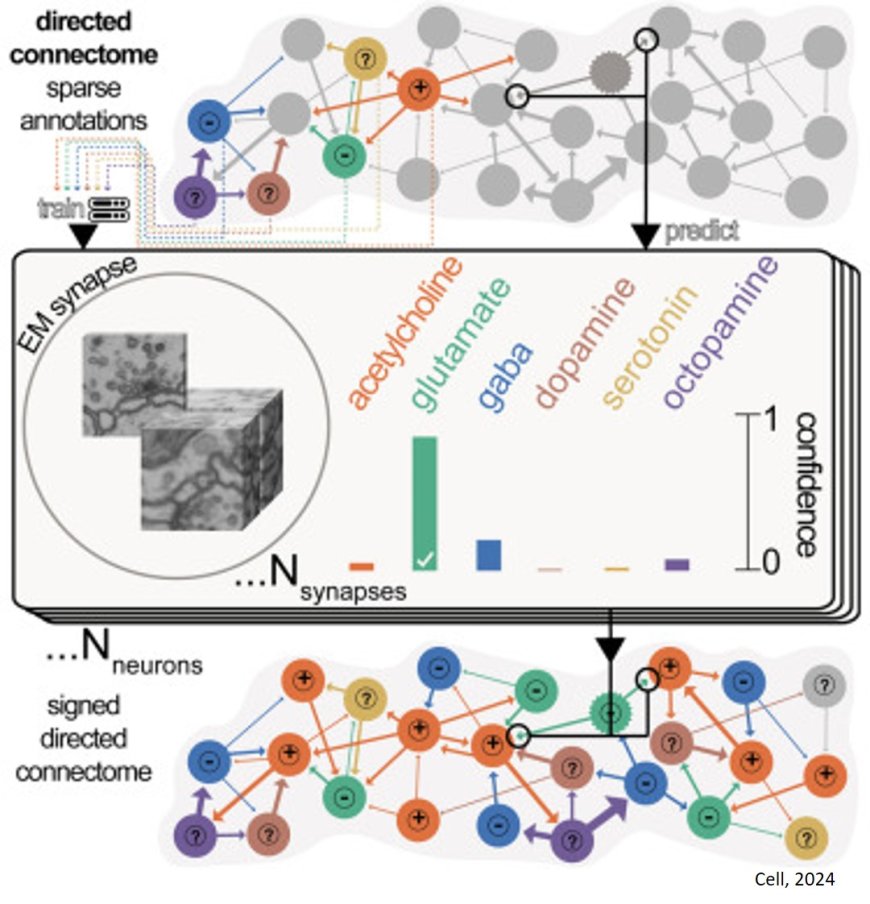
The group wondered if they might be able to use information from previous experiments identifying the neurotransmitters released from some neurons to predict the neurotransmitters released from others in the connectome. Neurons use neurotransmitters to communicate with each other, with different chemicals responsible for different signals.
The human eye can’t tell the difference between the synapses on neurons where different neurotransmitters are released, but perhaps a computer model could. Funke and his colleagues were skeptical, but they thought it might be worth giving it a try.
“This is basically where we left it: we have the data, I guess we could try,” the senior author says. “We were not particularly optimistic.”
The project was given to a high school student who was starting a summer internship in the lab. The project would allow the student to learn how to train a neural network to recognize images – a useful skill for a budding computer scientist even if the project did not yield results.
The author having trained the model on published data and evaluated its performance on test data. Though they had little hope it would work, the model was more than 90 percent accurate in predicting some neurotransmitters.
“I couldn’t believe it,” the senior author says. “The numbers were way too good.”
After checking the data and the model, the authors were convinced that the numbers weren’t a mistake: The model could predict neurotransmitters. But the team was still cautious, and they didn’t have a good grasp on how the network was making the predictions.
“I should have been very happy, but instead I was worried because we didn’t understand what was going on,” the author says.
After ruling out possible confounders that could be skewing their results, the team developed a way to understand what the network was seeing that allowed it to make predictions.
First, they used their network to predict a neurotransmitter from a known image, which it did successfully. Then, they asked a separate network to take that known image and change it slightly to create an image corresponding to the release of a different neurotransmitter – essentially identifying the minimum traits that need to be changed for the model to predict one neurotransmitter over another. Lastly, the team developed a separate method to identify these distinct traits.
From this information, the team understood the different features their original network used to make predictions. This gave them confidence to release their method to the wider neuroscience community in 2020.
“What most of the neuroscience community has seen from this work is the predictions,” the senior author says. “They were happy to use it, but for us it was very important to make sure it was actually working.”
Five years later, the student is now an undergraduate at Duke University, and the method has been used to predict neurotransmitters in connectomes of the fruit fly hemibrain, ventral nerve cord, and optic lobe created by the researchers and collaborators, as well as the adult fly brain connectome created by FlyWire.
The information helps scientists understand how neurons in a circuit affect each other so they can then form hypotheses about the function of brain circuits that can be tested in the lab.
“It really all started with a bit of a crazy idea, something that no one was really too optimistic about. And what do you do with a crazy idea? You give it to a high school student as a learning experience,” the author says.