AI used to create proteins that kill E. coli
In the last year, there has been a surge in proteins developed by AI that will eventually be used in the treatment of everything from snakebites to cancer. What would normally take decades for a scientist to create – a custom-made protein for a particular disease – can now be done in seconds.
For the first time, scientists have used Artificial Intelligence (AI) to generate a ready-to-use biological protein, in this case, one that can kill antibiotic resistant bacteria like E. coli.
This study, published in Nature Communications, provides a new way to combat the growing crisis caused by antibiotic resistant super bugs. Iron is growth-limiting during infection and bacterial pathogens encode transporters capable of extracting the iron-containing cofactor heme directly from host proteins.
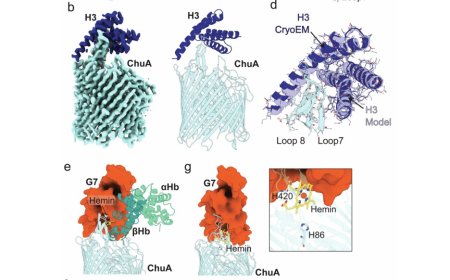
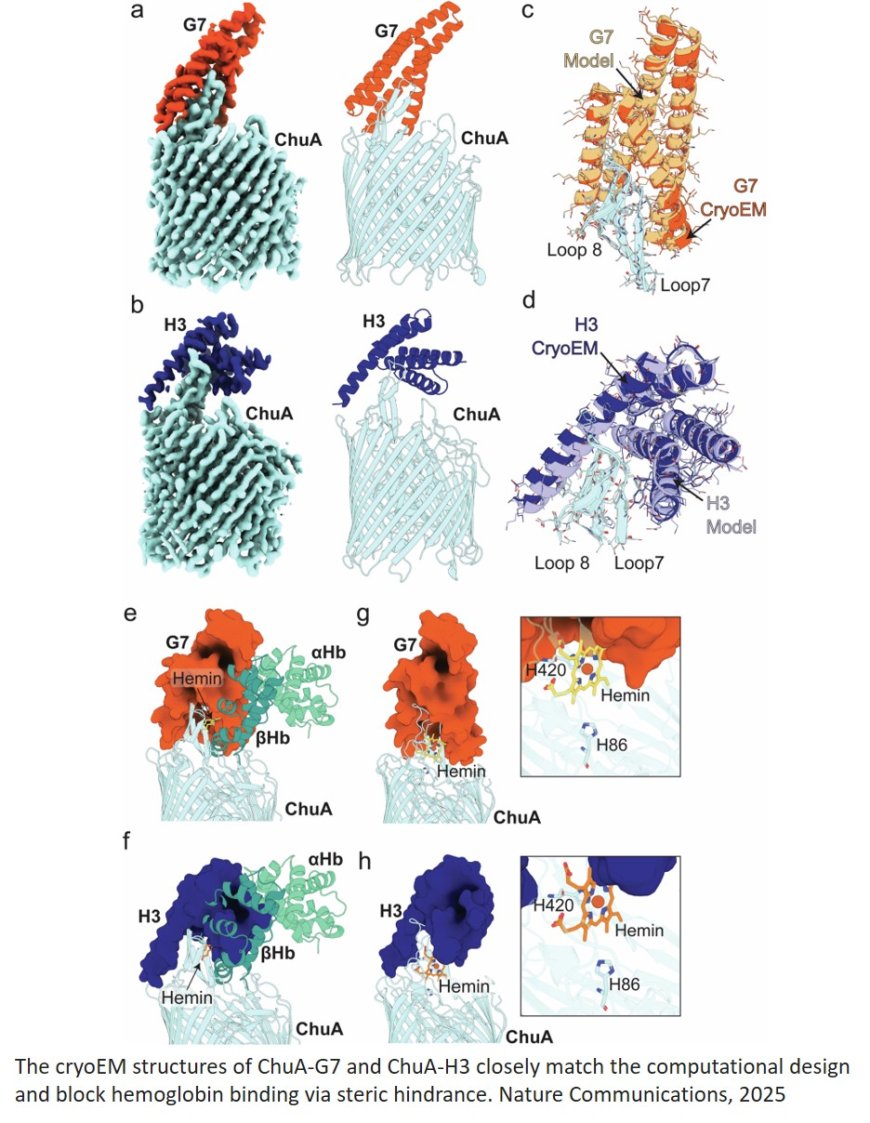
Bacterial pathogens encode transporters capable of extracting the iron-containing cofactor heme directly from host proteins. Pathogenic E. coli and Shigella spp. produce the outer membrane transporter ChuA, which binds host hemoglobin and extracts its heme cofactor, before importing heme into the cell. Heme extraction by ChuA is a dynamic process, with the transporter capable of rapidly extracting heme from hemoglobin in the absence of an external energy source, without forming a stable ChuA-hemoglobin complex.
By using AI in this way, scientists have developed AI platforms capable of rapidly generating thousands of ready-to-use proteins, paving the way for faster, more affordable drug development and diagnostics that could transform biomedical research and patient care.
They created binders capable of inhibiting E. coli growth by blocking hemoglobin binding to ChuA. The authors identified several binders that inhibit E. coli growth at low nanomolar concentrations, without experimental optimisation.
The authors determined the structure of a subset of these binders, alone and in complex with ChuA, demonstrating that they closely match the computational design.
The AI Protein Design Platform used in this study and AI-driven protein design tools are freely available for scientists everywhere. “It’s important to democratize protein design so that the whole world has the ability to leverage these tools,” said the lead author of the study. “Using these tools and those we are developing in-house, we can engineer proteins to bind a specific target site or ligand, as inhibitors, agonists or antagonists, or engineered enzymes with improved activity and stability.”
Currently proteins used in the treatment of diseases like cancer or infections are derived from nature and repurposed through rational design or in vitro evolution and selection. “These new methods in deep learning enable efficient de novo design of proteins with specific characteristics and functions, lowering the cost and accelerating the development of novel protein binders and engineered enzymes,” the author said.
https://www.nature.com/articles/s41467-025-60612-9
https://sciencemission.com/Inhibiting-heme-piracy-by-pathogenic-Escherichia-coli