Tool for classifying tumor structural variants
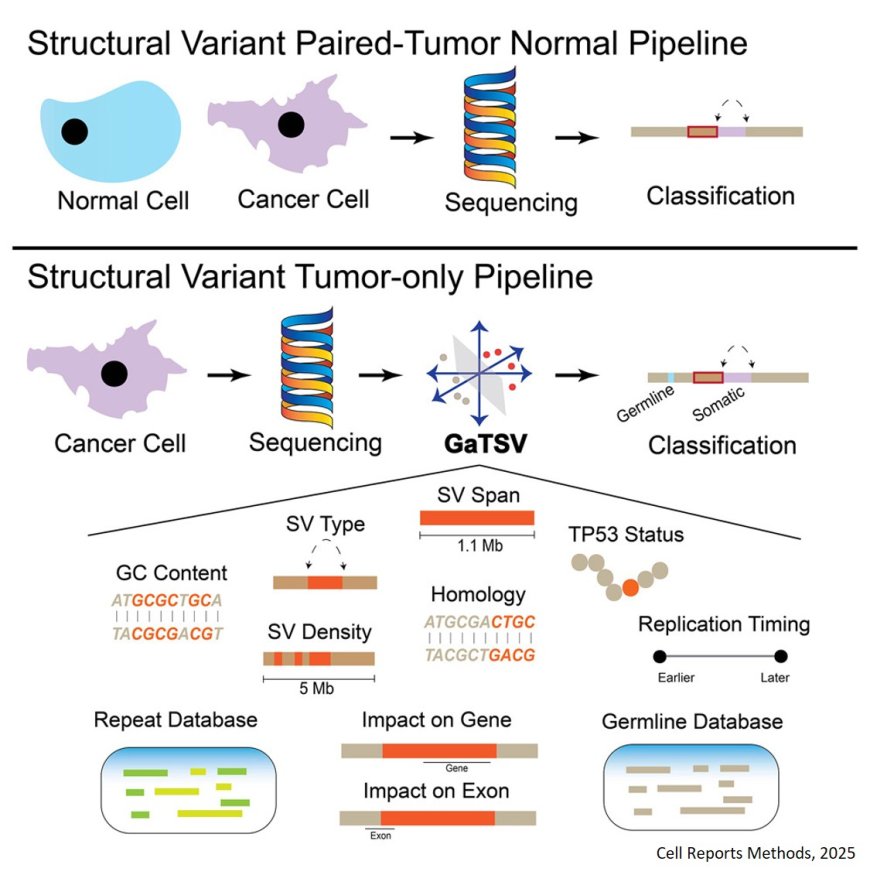
Genomic structural variants (SVs) are key to understanding cancer and can inform treatment response.
Distinguishing cancer from normal SVs has the limitation of requiring paired normal samples.
The researchers looked at similarities and differences between 2 million germline and 115 thousand tumor SVs from a cohort of 963 patients from The Cancer Genome Atlas.
They identified features linked to transposon-mediated processes that are associated with germline SVs, while somatic SVs more frequently show features characteristic of chromoanagenesis.
The results enabled the authors to introduce the -Germline and Tumor Structural Variant (GaTSV), a classifier that accurately identifies cancer-specific SVs without matched normal tissue, based on genomic sequence context differences.
https://www.cell.com/cell-reports-methods/fulltext/S2667-2375(25)00027-X
https://sciencemission.com/classifying-tumor-structural-variants